Research
The Department of Cell Biology was established in September 2012 and moved into the BMC in summer 2015. A long-term goal of the Kiebler lab is to understand the molecular basis of synaptic plasticity. Here, we are particularly interested in how individual synapses, the point of contact and communication between neurons, are altered during their lifetime and how this contributes to our ability to learn and remember. It has recently emerged that one important underlying mechanism is dendritic RNA localization and subsequent translational control at the synapse. In these crucial processes, RNA-binding proteins (RBPs) play essential roles. Over the last decade, the Kiebler lab has identified a series of RBPs, e.g. Staufen2, Pumilio2 and Barentsz and has extensively characterized their functional contribution to dendritic RNA localization and translational control at the synapse.
In the following, there is a short summary of current research projects that are currently performed in the lab.
1. RNA-binding proteins as functional regulators of non-coding RNAs at the synapse
(funded by the DFG, SPP1738 (http://www.spp1738.de)
Summary: In mature hippocampal neurons, a subset of RNAs localizes to dendrites within ribonucleoprotein particles (RNPs). These RNPs not only contain localized mRNAs and RNA-binding proteins (RBPs), e.g. the brain-specific double-stranded RNA-binding protein Staufen2 (Stau2), Barentsz (MLN51, Casc3) and Pumilio 2 amongst others, but also a set of non-coding mRNAs. Together with RBPs known to regulate translation, these miRNAs are crucial to keep mRNAs in a translationally repressed state during transport to the synapse. In this project, we aim to identify non-coding mRNAs in Stau2 and Pum2 RNPs and study the underlying mechanism of translational control occurring at individual synapses of hippocampal neurons. In conclusion, we hope to understand how local protein synthesis contributes to morphologically and functionally reorganize individual synapses upon learning and memory.
2. Pumilio2-mediated control of local protein expression in neurons
(funded by the DFG, http://for2333.de)
Summary: To elucidate spatially restricted translational control in mammalian neurons, we aim to identify physiological targets of the translational repressor Pumilio2 (Pum2) and how they are regulated by Pum2. Therefore, we plan to combine RNA granule isolation with in vitro translation assays in collaboration with Jan Medenbach (U Regensburg) and in cellulo assays using isolated neurons from Pum2-knock down mice that are available in our lab.
3. The role of Stau2 in neuronal RNPs
(funded by the DFG, http://for2333.de)
Summary: Here, we study neuronal mRNA transport in primary neurons by focusing on the double-stranded RNA-binding protein Staufen2. These experiments will be complimented by in vitro studies of the Niessing lab (link to https://www.helmholtz-muenchen.de/stb/research/groups/research-group-niessing/research/index.html)
4. The in vivo role of RNA-binding proteins in neurogenesis, synapse formation and synaptic plasticity
(funded by the DFG, http://www.sfb870.mcn.uni-muenchen.de/index.html)
Summary: RNA-binding proteins (RBPs) play key roles in the fate of mRNAs within a cell, e.g. the asymmetric segregation of mRNAs into specific subcellular compartments in polarized cells. This enables neuronal differentiation and synaptic function. In this context, the Kiebler lab has previously characterized two important RBPs, Staufen2 (Stau2) and Pumilio2 (Pum2) and their contribution to dendritic mRNA localization and local protein synthesis at the activated synapse.
In this project, we aim to unravel the role of Stau2 and Pum2 in asymmetric mRNA distribution in neuronal stem cells, neuronal progenitors and mature neurons in the hippocampus. Furthermore, we plan to evaluate their influence to neuronal differentiation, maturation and functional integration into existing neural circuits. We are taking advantage of two new transgenic rat models that allow either tissue-specific, Cre-dependent or constitutive silencing of Stau2 (Stau2KD), and gene-trap mice that are deficient for Stau2 (Stau2GT), Pum2 (Pum2GT) or both (Stau2GT-Pum2GT).
First, we will investigate the role of Stau2 and Pum2 in adult neurogenesis by (i) studying RBP-dependent asymmetric mRNA distribution in neuronal progenitors; (ii) determining the neurogenesis rate in WT vs. Stau2GT or Pum2GT adult hippocampus; and (iii) investigating learning-induced neurogenesis. Second, we will unravel the in vivo role of Stau2 and Pum2 in synaptic plasticity and therefore in learning and memory. Interestingly, the above-mentioned animal models, e.g. Stau2KD and Stau2GT both show deficits in hippocampal synaptic plasticity and in hippocampal spatial working memory, novelty detection and temporal association memory. We will investigate whether there are alterations in the asymmetric distribution of Stau2 mRNA targets encoding synaptic proteins, both in cultured Stau2KD rat hippocampal neurons as well as in brain sections derived from Stau2KD rats. Furthermore, we will study synaptic activity-dependent mRNA localization and translation of these Stau2 targets both in vitro and in vivo.
We are convinced that dissecting the molecular mechanisms of how Stau2 and Pum2 contribute to hippocampal neurogenesis, synaptic plasticity and working memory will advance our understanding of remarkably diverse cognitive and neurological symptoms both during healthy ageing as well as in many neurological diseases.
5. Imaging the fate of Rgs4 mRNA in living neurons using the modified MS2 system
(collaboration with the Bertrand lab in Montpellier, France
(http://www.igmm.cnrs.fr/en/team/biogenese-des-arns)
previously funded by the FWF, Austria and ANR, France)
Summary: In order to obtain a more detailed understanding of how RBPs like Staufen proteins contribute to these processes, we are choosing a live cell imaging approach in living neurons and are analyzing the assembly and transport dynamics of individual RNP granules. We have therefore established an improved version of the MS2 RNA imaging system (Bertrand et al., 1998) with the help of Edouard Bertrand (IGMM, Montpellier, France) In brief, the MS2 system utilizes the specific, high affinity interaction between a 19-nucleotide stem-loop structure of the MS2 phage RNA and the MS2 coat protein (MCP) (Figure 1).
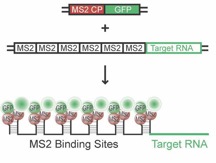
Figure 1: The MS2 RNA imaging system consisting of two components (Bertrand et al., 1998; cartoon from: Jambhekar & Derisi, 2007). GFP-tagged MS2 coat protein (MS2 CP = MCP, plasmid 1) binds the MS2-repeats fused to the target mRNA (plasmid 2) with high affinity, allowing for visualization of individual RNA by live cell imaging in primary neurons.
Co-expressing the fluorescently tagged MS2 coat protein (MCP), together with a reporter RNA of interest fused to consecutive repeats of the MS2 stem-loop motif, allows us to study the localization and the dynamics of Rgs4 mRNA by live cell imaging in high spatiotemporal resolution. These live cell imaging experiments show that Rgs4 mRNA is delivered in RNA granules into dendrites of living primary neurons and that the observed transport occurs bidirectionally. Interestingly, we detect a slight but significant bias of Rgs4 mRNA transport towards the anterograde direction. In conclusion, this approach will not only allow us to determine the role of possible cis-acting signals and their role in mRNA transport, but also to investigate RNP assembly.
6. Determining Staufen2 binding of its physiological mRNA targets using iCLIP
(collaboration with the Ule lab in London, UK
(https://www.crick.ac.uk/research/a-zresearchers/researchers-t-u/jernej-ule)
previously funded by HFSP, France)
Summary: To gain more detailed insight into how Stau2 recognizes its target, we are collaborating with Jernej Ule (The Crick Institute & UCL, London, UK) to identify Stau2-RNA interactions in a global manner using UV-crosslinking and immunoprecipitation (CLIP). The Ule lab has recently developed the individual resolution CLIP method (iCLIP), which now allows to quantitatively determine the positioning of hnRNP particles on nascent transcripts with nucleotide resolution (König et al, 2010). It was a particularly demanding task to adapt iCLIP to the double-stranded RBP (dsRBP) Staufen, as it does not cross-link to its target RNAs with high efficiency (Sugimoto et al., 2015). We now have first high-coverage iCLIP data for Rgs4 mRNA with a strong preference for selective structured regions in its 3’-UTR: the top two cross-link sites are close to SRS elements.

